exosomes correlated with the genotype of the animal, with progressively less protein in the carrier and affected animals compared to wildtype mice. SMN protein was also easily detectable in exosomes isolated from human serum, with a reduction in the amount of SMN protein contained in exosomes from patients with SMA compared to normal controls. We are currently testing serum-derived exosomes from patients with SMA who are receiving treatment for the disease, to determine if this assay can be used to monitor response to therapy, to ultimately personalize the dose or timing of treatment for each patient. We are also performing proteomic analysis of serum-derived exosomes from patients with SMA and amyotrophic lateral sclerosis (ALS) to identify novel biomarkers of the disease state.
Nash, L.A., J.K. Burns, J. Warman Chardon, R. Kothary and R.J. Parks (2016) Spinal Muscular Atrophy: More than a disease of motor neurons? Current Molecular Medicine 16(9):779-792.
Nash, L.A, E.R. McFall, A.M. Perozzo, M. Turner, K.L. Poulin, Y. De Repentigny, J.K. Burns, H.J. McMillan, J. Warman Chardon, D. Burger, R. Kothary, and R.J. Parks (2017) Survival Motor Neuron Protein is Released from Cells in Exosomes: A Potential Biomarker for Spinal Muscular Atrophy. Scientific Reports 7(1):13859.
Development of improved oncolytic virus for cancer therapy.
Although oncolytic vectors, which are designed to specifically replicate in and kill cancer cells, have shown great promise in preclinical studies, they have shown more limited efficacy in vivo partly due to an inability to efficiently spread through the tumor mass. We are investigating whether oncolytic adenovirus (Ad) vectors expressing a fusogenic protein, which can mediate cell-cell fusion, can enhance spread through the tumor.
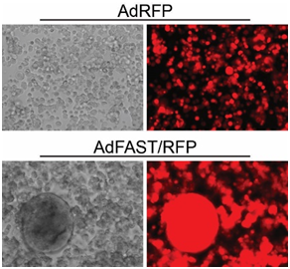
We are evaluating replication-defective and replication-competent (oncolytic) Ad vectors expressing the p14 Fusion Associated Small Transmembrane (FAST) protein in tissue culture and animal models of cancer. In the tissue culture models, these vectors perform extremely well and mediate efficient fusion of the cells into a multi-nucleated syncytium. In mouse models of cancer, the replication-competent oncolytic Ad expressing p14 FAST shows superior efficacy relative to vector lacking the fusogenic protein. We are continuing our studies in other mouse models of cancer, to optimize the design and efficacy of the vectors.
Del Papa, J. and R.J. Parks (2017) Adenoviral Vectors Armed with Cell Fusion-Inducing Proteins as Anti-Cancer Agents. Viruses 19;9(1). pii: E13. doi: 10.3390/v9010013.Wong, C.M., K.L. Poulin, G. Tong, C. Christou, M.A. Kennedy, T. Falls, J.C. Bell and R.J. Parks (2016) Adenovirus-mediated expression of the p14 fusion-associated small transmembrane protein promotes cancer cell fusion and apoptosis in vitro but does not provide therapeutic efficacy in a xenograft mouse model of cancer PLoS ONE 11(3):e0151516. doi: 10.1371/journal.pone.0151516.
Epigenetic regulation of the adenovirus genome.
Ad-based vectors can persist in a transduced cell in mouse liver for the lifetime of the animal (~2 years), or up to 7 years in non-human primates. However, little is known about the structure of the virus in the infected cell nucleus. We have performed a number of studies to examine the nucleoprotein structure of Ad DNA within
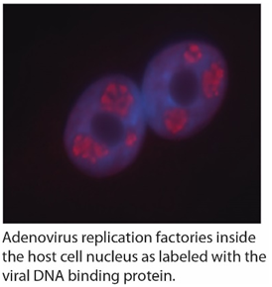
the host cell nucleus. We showed that both Ad gene therapy vectors and wildtype virus associate with histones and are wrapped in a chromatin-like structure similar to the host cell genome. This event appears to be important for optimal expression of virus encoded genes. We have gone on to identify some of the cellular proteins involved in Ad “chromatinization”. We have also performed high throughput screens to identify small molecules that impact virus replication, including testing a panel of drugs that specific target epigenetic regulatory proteins within the cell. Elucidating the mechanism of how these drugs and cellular proteins affect the virus will provide novel insight into Ad replication and its function as a gene therapy vector.
Wong, C.M., E.R. McFall, J.K. Burns and R.J. Parks (2013) The role of chromatin in adenoviral vector function. Viruses 5:1500-1515.
Giberson, A.N., B. Saha, K. Campbell, C. Christou, K.L. Poulin, and R.J. Parks. (2018) Human adenoviral DNA association with nucleosomes containing histone variant H3.3 during the early phase of infection is not dependent on viral transcription or replication. Biochem. and Cell. Biol. (in press).
